We are moving our newsletter to Substack for a better experience!
In Week #244 of the Doctor Penguin newsletter, the following papers caught our attention:
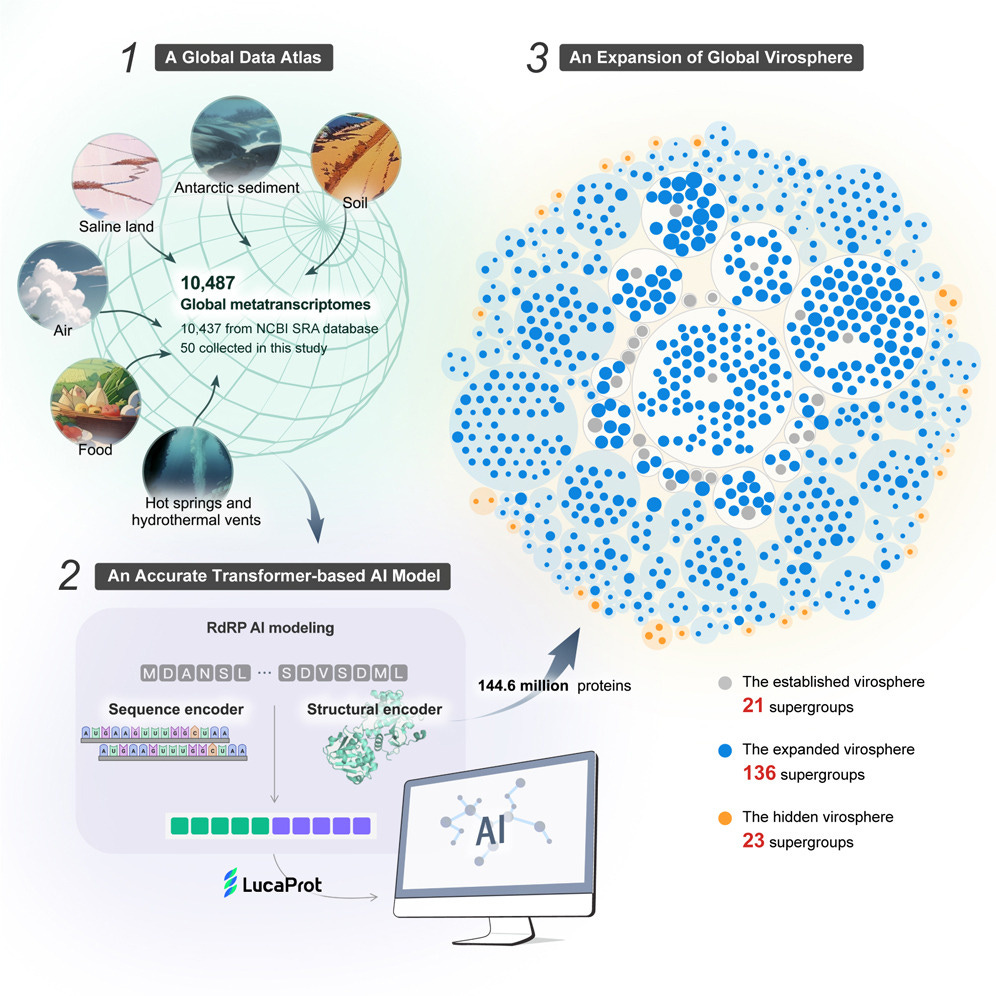
1. RNA Virus Discovery. Large-scale virus discovery studies rely on analyzing RNA-dependent RNA polymerase (RdRP) sequences, a key component of RNA virus genomes. However, current metagenomic tools often miss highly divergent RdRPs, suggesting that many novel RNA virus groups remain undiscovered, hindering advancements in RNA virus evolution and ecology.
Hou et al. developed LucaProt, a transformer-based model that integrates protein sequence composition and structure information to identify highly divergent viral RdRPs. Trained on 235,413 samples (5,979 known viral RdRPs and 229,434 non-viral RdRP proteins), this method outperforms conventional approaches in accuracy, efficiency, and breadth of virus diversity detected. By analyzing over 10,000 metatranscriptomes from diverse global ecosystems, LucaProt identified 161,979 potential RNA virus species and 180 RNA virus supergroups, including many previously understudied groups. This represents a major expansion of known RNA virus diversity, with newly discovered viruses present in diverse environments such as air, hot springs, and hydrothermal vents. The study confirmed the RNA nature of some virus supergroups through simultaneous RNA and DNA sequencing of the same samples, and it revealed exceptionally long RNA virus genomes and complex genomic structures. While the hosts for most identified viruses remain unknown, this study significantly advances our understanding of the global RNA virome's scale and diversity, providing new computational tools for future virus discovery efforts.
Read paper | Cell
2. Autism. Early autism detection is crucial for timely intervention and improved outcomes, but diagnoses are often delayed despite clinicians' ability to reliably identify autism in toddlers.
Babu et al. evaluated SenseToKnow, a mobile app for remote autism screening in toddlers aged 16-40 months. The app, downloadable on iPhones or iPads, displays movies and a game while recording children's behavioral responses via the device's camera and touch sensors. It uses computer vision models to extract behavioral features from video recordings, which are then age-adjusted and fed into an XGBoost model for autism classification. The study included 620 toddlers, with 188 subsequently diagnosed with autism by expert clinicians. Results demonstrated high diagnostic accuracy: AUROC of 0.92, sensitivity of 83.0%, and specificity of 93.3%. Performance was consistent across devices, genders, and diverse racial and ethnic backgrounds. This approach could reduce barriers to early autism screening, particularly benefiting families facing geographical, scheduling, or financial obstacles to clinic visits.
Read Paper | NEJM AI
3. Sensor Data. A lightweight model for processing medical time series data.
Chen et al. developed SMoLK, a lightweight and interpretable architecture for medical time series processing. SMoLK is essentially equivalent to a linear combination of filtered signals generated by convolving the input with a set of learnable kernels, which allows direct computation of each input signal's contribution to the output by reversing the convolution process. SMoLK achieved state-of-the-art performance in PPG artifact segmentation with far fewer parameters than current models, and matched deep ResNet performance in ECG classification using less than 1% of the parameters. The largest model requires under 100 KB of memory (75 KB after pruning), while the smallest needs only 3 KB, making it suitable for real-time processing in wearable devices. This efficiency allows SMoLK to perform signal quality assessment or arrhythmia classification as a background process in wearables.
Read Paper | Nature Machine Intelligence
-- Emma Chen, Pranav Rajpurkar & Eric Topol