Week 235
Dementia, LLM, EHR Foundation Model, Impairment quantification, Organoids & Organ-on-a-Chip
We are moving our newsletter to Substack for a better experience!
In Week #235 of the Doctor Penguin newsletter, the following papers caught our attention:
1. Dementia. Dementia is not a specific disease, but rather a general term for the impaired ability to remember, think, or make decisions that interferes with performing everyday activities. Differentiating between the various types of dementia remains a challenge in neurology, primarily due to the overlapping symptoms of different dementia etiologies.
Xue et al. developed a transformer-based model for differential diagnosis of dementia using multimodal data, including demographics, personal and family medical history, neuropsychological assessments, functional evaluations, and multimodal neuroimaging. The model addresses the complex clinical challenge of distinguishing between various dementia etiologies, including Alzheimer’s disease, vascular dementia, and Lewy body dementia. It provides probability scores for cognitive status (normal cognition, mild cognitive impairment, dementia) and 10 distinct dementia etiologies, maintaining reliability even with missing data by incorporating random feature masking during training. Developed and validated on over 51,000 participants across 9 diverse datasets, the model achieved high performance in classifying cognitive states (microaveraged AUROC of 0.94) and differentiating dementia etiologies (microaveraged AUROC of 0.96). The model's predictions were corroborated by biomarker and postmortem data. Furthermore, in a randomly selected subset of 100 cases, the AUROC of neurologist assessments augmented by the model exceeded neurologist-only evaluations by 26.25%. These results demonstrate the model’s significant potential to be integrated as a screening tool for dementia in clinical settings and drug trials.
Read paper | Nature Medicine
2. LLM. Are current large language models (LLMs) ready for autonomous clinical decision-making in realistic medical scenarios?
Hager et al. conducted a comprehensive evaluation of LLMs for clinical decision-making using 2,400 real patient cases covering four common abdominal pathologies from the MIMIC-IV dataset. They assessed LLMs beyond just diagnostic accuracy, examining their ability to gather information, adhere to guidelines, interpret lab results, follow instructions, and maintain robustness to changes in input. During the experiment, they provided LLMs with a patient's history of present illness and asked them to iteratively gather and synthesize additional information such as physical examinations, laboratory results, and imaging reports until they were confident enough to provide a diagnosis and treatment plan. They found that LLMs performed worse than human clinicians in diagnostic accuracy, especially when required to gather information themselves. LLMs often failed to order necessary exams, tended to diagnose prematurely, and struggled with interpreting lab results even when provided with reference ranges. Counterintuitively, LLMs were sensitive to the order in which information was presented, resulting in large changes in diagnostic accuracy despite identical diagnostic information. They also performed worse when all diagnostic exams were provided, typically attaining their best performance when only a single exam was provided in addition to the history of present illness. Besides, they failed to consistently recommend appropriate and sufficient treatments, particularly for severe cases. Finally, LLMs were sensitive to small changes in instructions and information presentation, leading to significant variations in diagnostic accuracy. While some improvements were achieved through summarization techniques and removing normal lab results, the study shows that current LLMs are not yet suitable for autonomous clinical decision-making due to their limitations in accuracy, consistency, and robustness.
Read Paper | Nature Medicine
3. EHR Foundation Model. Is it feasible to share foundation models across hospitals as a less expensive route for local hospitals to adapt a foundation model for their specific needs?
Guo et al. conducted a multi-center study to evaluate the adaptability of CLMBR-T-base, a publicly available foundation model, across different hospital systems. This model was pretrained on longitudinal, structured medical records from 2.57 million Stanford Medicine patients and tested on datasets sourced from The Hospital for Sick Children in Toronto and Beth Israel Deaconess Medical Center in Boston. The study assessed the model's adaptability through continued pretraining on local data and compared its performance on 8 clinical prediction tasks against baseline gradient boosting machines (GBMs) as well as locally trained foundation models. Results showed that the adapted external foundation model matched or exceeded GBMs trained on all available local data, while requiring an average of just 128 training examples (less than 1% of the total data) to match the baseline performance of GBMs trained on all available task data. Notably, continued pretraining produced foundation models similar in performance to those trained from scratch, but required 60 to 90% less patient data for pretraining, demonstrating significant improvements in data efficiency and adaptability across diverse healthcare settings.
Read Paper | npj Digital Medicine
4. Impairment quantification. Develop data-driven quantitative metrics of impairment and disease severity using data from healthy subjects only.
Yu et al. developed a framework called COBRA for quantifying impairment and disease severity using models trained exclusively on healthy individuals. The key idea is to exploit the decrease in model confidence when the models are applied to data from impaired or diseased patients, which indicates a deviation from the healthy population. The framework was demonstrated in two applications: quantifying stroke-induced upper-body impairment and assessing knee osteoarthritis severity. For stroke patients, convolutional neural networks were trained on wearable sensor and video data from healthy individuals to predict functional primitives. The COBRA score, computed as the average model confidence for motion-related primitive predictions (transport, reposition, and reach), showed a strong correlation with the gold-standard Fugl-Meyer Assessment. For knee osteoarthritis, a Multi-Planar U-Net model was trained to perform tissue segmentation in 3D MRI scans of healthy individuals, with the COBRA score computed from the confidence for voxels identified by the model as cartilage. This score is again highly correlated with the Kellgren-Lawrence grading system. Although this study highlights the potential of the COBRA framework to enable more frequent, objective patient monitoring, the authors caution that extraneous factors may produce a spurious decrease in the confidence of the AI model.
Read Paper | npj Digital Medicine
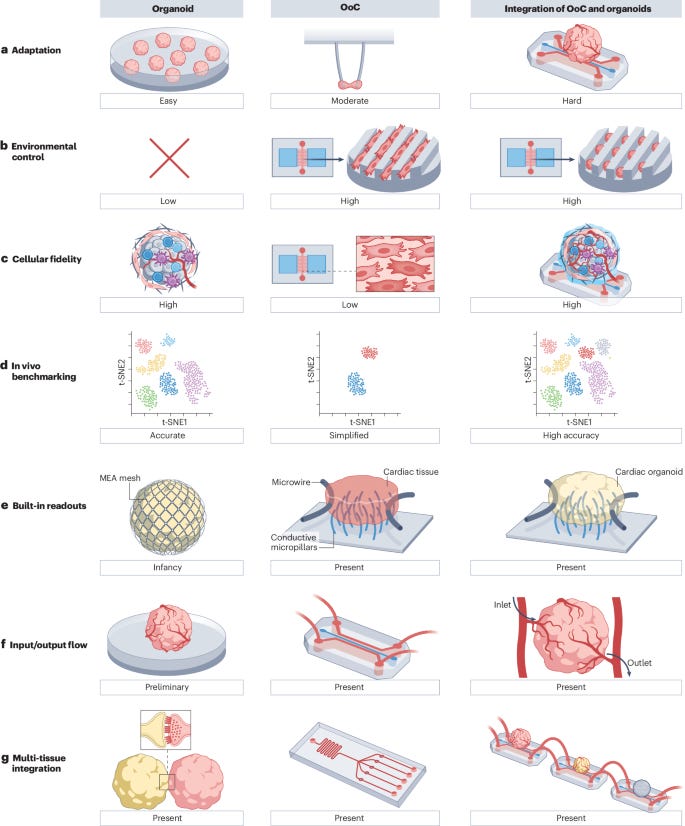
5. Organoids & Organ-on-a-Chip. Organoids and organs-on-chips (OoCs) are two rapidly emerging 3D cell culture techniques that aim to bridge the gap between in vitro 2D cultures and animal models to enable clinically relevant drug discovery and model human diseases. In 3D cultures, cells are surrounded by other cells and the extracellular matrix, mimicking their arrangement in the body. This contrasts with 2D cultures, where cells grow on flat plastic or glass surfaces. Organoids are self-organized 3D tissue structures derived from stem cells that mimic organ development. OoCs are engineered microfluidic devices that recreate tissue-specific microenvironments to derive miniature functional tissue models with desired tissue organization by combining cells, scaffolds, and topographical guidance.
In this review, Zhao et al. explore the integration of organoids and OoCs technologies to create more physiologically relevant in vitro models for drug development and disease research. They highlight how this combination leverages the strengths of both approaches: organoids provide cellular complexity and self-organization, while OoCs offer precise environmental control and the ability to incorporate various stimuli (such as electrical, mechanical, and shear stress) and sensors (like oxygen probes). This integration addresses key challenges in both fields, including tissue vascularization, creating high-fidelity parenchymal-vascular interfaces with multiple cell types, enabling multi-organ communication through connected chips, and incorporating sensors and stimuli for tissue maturation and real-time functional assessment. The review notes that despite the accelerated identification of promising molecules through AI, testing these molecules in animal models still results in a slow drug discovery cycle. The next frontier may lie in combining OoCs or organoids with AI approaches. By combining the high cellular fidelity of organoids with the controlled microenvironment of OoCs, these integrated systems have the potential to better recapitulate native tissue function, potentially revolutionizing drug discovery and disease modeling.
Read Paper | Nature Reviews Bioengineering
-- Emma Chen, Pranav Rajpurkar & Eric Topol