We are moving our newsletter to Substack for a better experience!
In Week #232 of the Doctor Penguin newsletter, the following papers caught our attention:
1. Antibiotics Discovery. Unveil molecules that may play a role in host immunity throughout evolution by discovering antimicrobial peptides from extinct organisms.
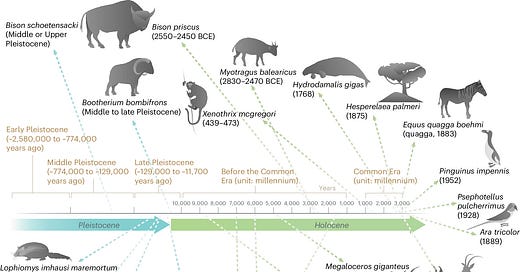
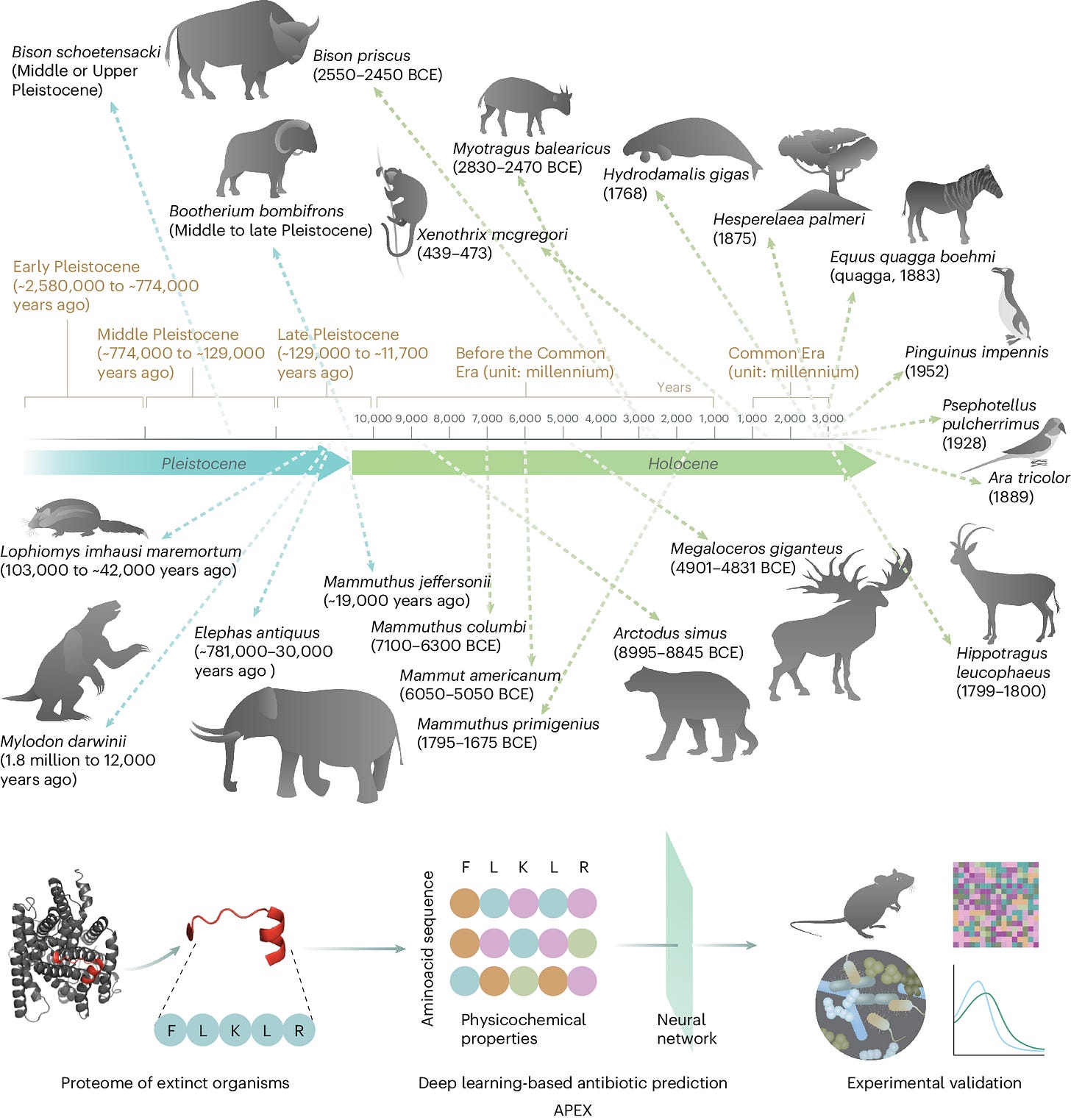
Wan et al. developed APEX, a multitask deep learning approach for discovering new antimicrobial peptides by mining the proteomes of extinct organisms. APEX employs an encoder neural network that combines recurrent and attention neural networks to extract hidden features from peptide sequences. These features are then used to predict the antimicrobial activity of each input peptide against specific bacterial strains (regression output) and to classify each peptide as antimicrobial or non-antimicrobial (binary classification output). APEX outperformed previous computational methods and identified 37,176 peptides predicted to have broad-spectrum antimicrobial activity, including 11,035 unique to extinct organisms. Experimental validation of 69 of these peptides revealed potent activity against clinically relevant bacterial pathogens. Notably, lead peptides derived from the woolly mammoth, straight-tusked elephant, ancient sea cow, giant sloth, and extinct giant elk demonstrated anti-infective efficacy in mouse models of skin and thigh infections, comparable to a clinical antibiotic.
Read paper | Nature Biomedical Engineering
2. LLM. Backpropagation via text.
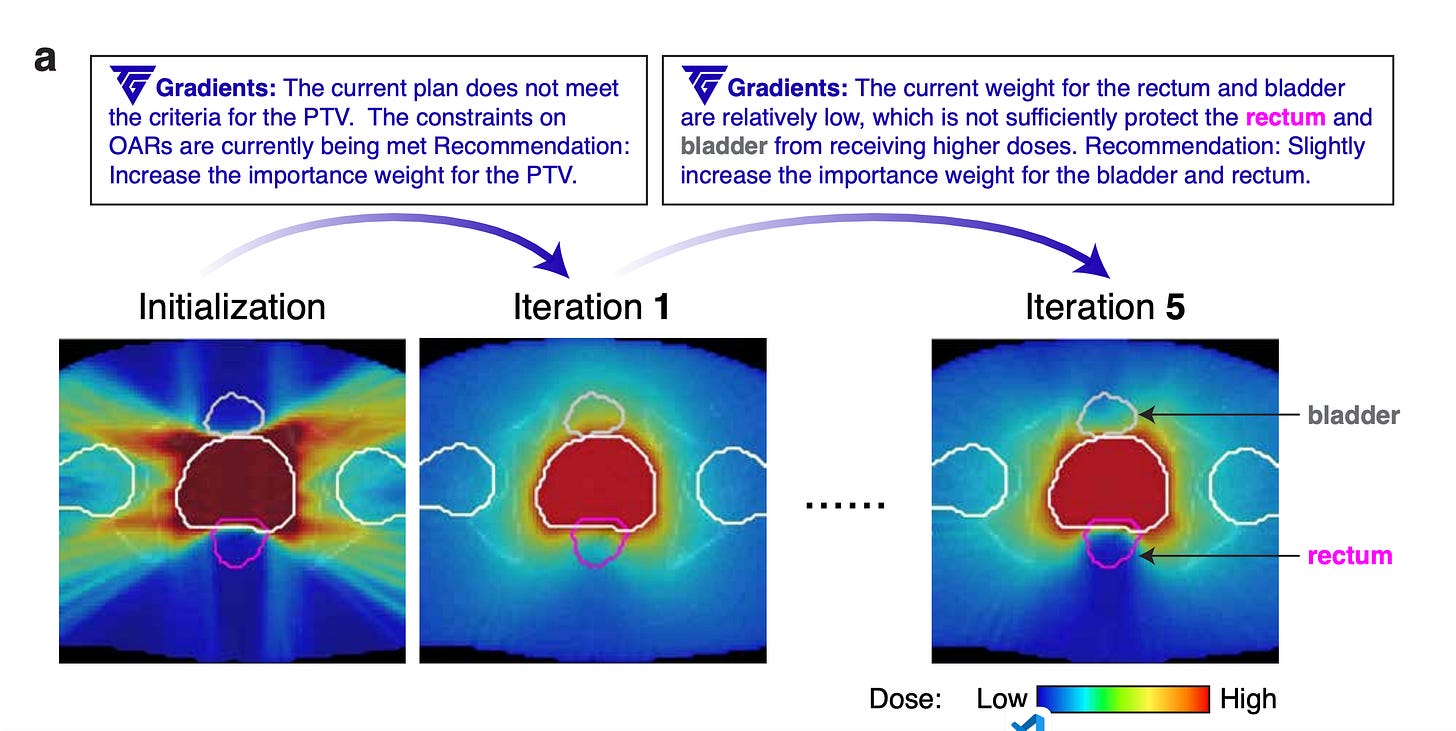
Inspired by the success of backpropagation and automatic differentiation in training neural networks, Yuksekgonul et al. developed TEXTGRAD, a framework that performs automatic "differentiation" via text to optimize individual components of compound AI systems, where each component could be an LLM-based agent, a tool such as a simulator, or web search. The framework first transforms each AI system into a computation graph, where variables are inputs and outputs of complex (not necessarily differentiable) function calls. LLMs then generate informative and interpretable natural language criticism (dubbed 'textual gradients') for (1) instance optimization, such as improving a code snippet at test time; or for (2) prompt optimization, which finds a prompt that enhances an LLM's performance across multiple queries for a given task. The authors showcase biomedical use cases for TEXTGRAD in designing druglike small molecules with desirable properties and optimizing radiation oncology treatment plans. Following PyTorch's syntax and abstraction, TEXTGRAD is flexible, easy to use across various tasks without requiring component or prompt tuning.
Read Paper | arXiv preprint
3. LLM Hallucination. Radiology report generation requires vision-language models (VLMs) to identify all diseases or other features of interest in a radiology image and generate a short report. Sometimes VLMs generate hallucinatory statements about prior exams despite not having access to any. How can we reduce VLMs’ hallucinations of prior exams while maintaining the clinical accuracy of the generated reports?
Banerjee et al. propose a simple fine-tuning method inspired by direct preference optimization (DPO) for suppressing VLMs from generating hallucinated prior exams in chest X-ray (CXR) reports. DPO is a technique that aligns the behavior of a pretrained language model with human preferences by fine-tuning the model on a dataset of preferred and dispreferred responses and penalizing dispreferred responses in the loss function. The authors identify reports that reference prior exams in MIMIC-CXR as the dispreferred dataset and create a preference dataset by using GPT-4 to remove references to prior exams in the original reports. After fine-tuning, the best-performing model achieves a 3.2-4.8x reduction in lines hallucinating prior exams compared to the pretrained model while maintaining similar clinical accuracy as measured by RadCliQ-V1 and RadGraph-F1 metrics. This work demonstrates that DPO can effectively suppress specific unwanted behaviors in medical VLMs in a data and compute-efficient manner.
Read Paper | arXiv preprint
4. Long-Tailed Disease Diagnosis. Can AI diagnose rare diseases better than human experts?
Agarwal et al. compared the performance of 227 human radiologists and two AI algorithms, CheXpert and CheXzero, in diagnosing chest pathologies from X-ray images, with a focus on the "long tail" of rare diseases. CheXpert is a traditional supervised learning algorithm that can diagnose 12 chest pathologies, while CheXzero is a zero-shot learning algorithm that can diagnose any chest pathology. The study found that CheXzero outperforms human radiologists across all prevalence levels, with humans performing significantly worse than CheXzero in the low prevalence bin and only marginally better than a random classifier. Interestingly, human performance was only weakly correlated with CheXzero performance, suggesting that the two focus on different features of an X-ray. Although CheXpert beats humans and matches CheXzero for the pathologies it can predict, its limited scope hinders its overall performance. This study suggests that zero-shot learning algorithms like CheXzero are catching up to or even surpassing humans in diagnosing rare diseases in the long tail.
Read Paper | AEA Papers and Proceedings
-- Emma Chen, Pranav Rajpurkar & Eric Topol