We are moving our newsletter to Substack for a better experience!
In Week #189 of the Doctor Penguin newsletter, the following papers caught our attention:
1. Parkinson’s Disease. Parkinson's disease is clinically heterogeneous, likely driven by different cellular mechanisms in each patient. However, current approaches cannot define the molecular heterogeneity to understand the mechanisms that drive the phenotypic subtypes.
D'Sa et al. developed machine learning models to predict the presence of Parkinson's disease and its subtypes from patient-derived neurons. Using stem cell technology, they generated healthy control neurons and neurons representing 4 Parkinson's subtypes via chemical induction or genetic mutation. Multidimensional fluorescence imaging then quantified organelle features in the cells. The images and the quantitative single-cell fluorescence features were used to train deep learing models to predict the disease and subtypes with high accuracy. The models identified mitochondria (organelles that generate energy) and lysosomes (organelles containing digestive enzymes) as key differentiators, confirming their biological importance in Parkinson's disease. This study shows machine learning applied to patient-derived cells is highly accurate at predicting disease subtypes, providing proof-of-concept for enabling mechanistic stratification and precision medicine in the future.
Nature Machine Intelligence
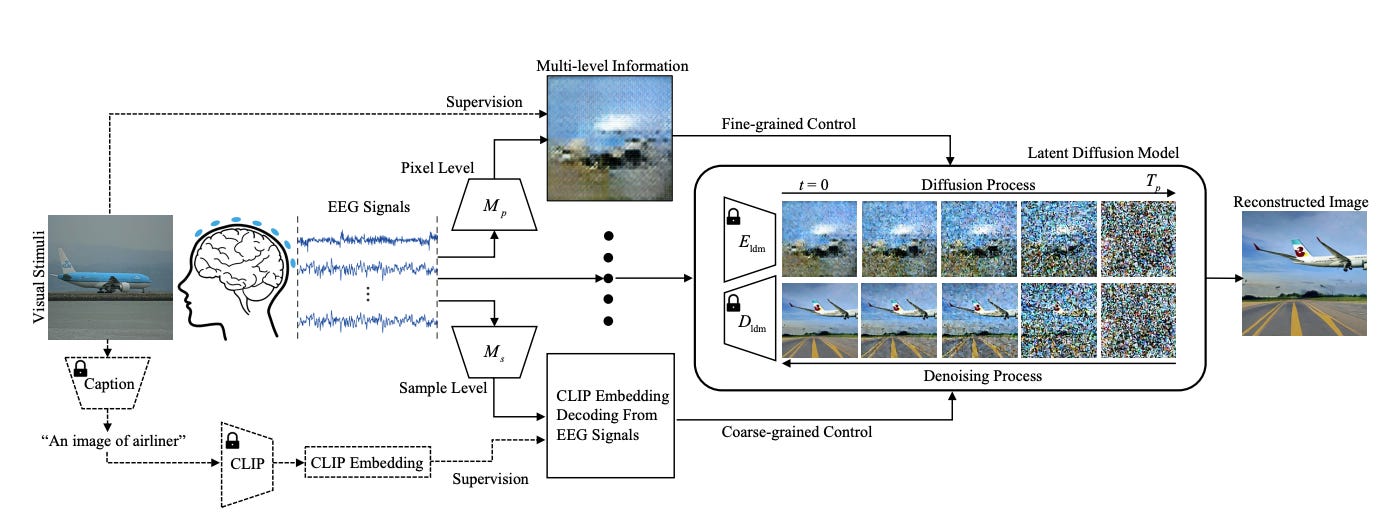
2. Visual Stimuli Reconstruction. Reconstructing images from brain activity can provide insights into the mechanisms of human visual perception and cognition.
Lan et al. proposed NEUROIMAGEN, a comprehensive pipeline to reconstruct visual stimuli images from electroencephalography (EEG) signals. NEUROIMAGEN first extracts multi-level semantic information from the EEG data, including pixel-level saliency maps and sample-level text descriptions. The multi-level semantic representations are then fused using a diffusion model to generate high-resolution reconstructed images. Specifically, saliency maps are generated using contrastive learning to align EEG embeddings from similar visual stimuli. A GAN model then produces the maps from the EEG features. Meanwhile, sample-level text descriptions are derived by aligning EEG signals to generated captions describing potential image content. This captures coarse-grained semantic information about the main objects in the image content. Though less detailed, these textual features provide accurate high-level information to aid reconstruction. Both qualitative and quantitative experiments demonstrate NEUROIMAGEN's capability in accurately recounstructing images from EEG data.
arXiv preprint
3. Histopathology. Proteomics involves the large-scale study of proteins, including their structures and functional roles. While rich proteomic data exists, there have yet to be prior attempts to correlate this information with histologic features from pathology imaging.
Wang et al. developed end-to-end, multi-resolution neural networks that combine proteomics, transcriptomics, and pathology imaging to identify predictive histology features associated with critical clinical outcomes in cancer. Trained on multi-modal data from 2,755 histology slides across 6 cancer types in the CPTAC dataset, the models effectively learned key pathological distinctions like tumor vs normal and tissue of origin. Incorporating the proteomic and transcriptomic data enabled identifying relevant biological pathways and cellular processes linked to the predictive histology features. Model generalizability and interpretability was confirmed using The Cancer Genome Atlas (TCGA). A clinical user interface was also developed to allow pathologists to apply these machine learning models to their own histological images.
Cell Reports Medicine
4. GPT-4. GPT-4 could suggest diagnoses in complex patients.
Shea et al. evaluated the diagnostic accuracy of GPT-4 versus clinicians in 6 elderly patients with delayed diagnosis over 1 month. The full medical histories were entered chronologically into GPT-4 without the final diagnosis. GPT-4's responses were compared to those of clinicians and a diagnostic decision support system. GPT-4 exceeded both clinicians and the system in primary and differential diagnosis accuracy. The study found that GPT-4 could suggest diagnoses not considered by clinicians before definitive investigations. Overall, GPT-4 showed potential for aiding diagnosis in complex older patients with comprehensive entries of demographic and clinical (including radiological and pharmacological) information, which may increase clinician confidence and prompt consideration of missing diagnoses.
JAMA Network Open
-- Emma Chen, Pranav Rajpurkar & Eric Topol