Week 188
Cancer of Unknown Primary, Neurodevelopmental Disorder, Cancer Screening, Medical Education
We are moving our newsletter to Substack for a better experience!
In Week #188 of the Doctor Penguin newsletter, the following papers caught our attention:
1. Cancer of Unknown Primary. Accounting for 3-5% of all cancers worldwide, cancer of unknown primary (CUP) cannot be traced back to its primary site, which can impact treatment choices due to differing clinical responses based on tumor types.
Moon et al. developed OncoNPC, a molecular cancer-type classifier trained on multicenter, targeted panel sequencing data from 36,445 tumors spanning 22 cancer types. OncoNPC predicted primary cancer types with high confidence in 41.2% of 971 CUP tumors from the Dana-Farber Cancer Institute. Moreover, OncoNPC was able to identify CUP subgroups with significantly higher polygenic germline risk for the predicted cancer types and with significantly different survival outcomes. In the retrospective survival analysis, CUP patients treated in line with OncoNPC's predictions had a notably longer survival rate. Finally, OncoNPC predictions enabled a 2.2-fold increase in patients with CUP who could have received genomically guided therapies. These findings suggest that CUP tumors share a genetic and prognostic architecture with known cancer types and may benefit from molecular classification.
Nature Medicine
2. Neurodevelopmental Disorder. Facial recognition is valuable in identifying (neuro) developmental syndromes due to the intrinsic connection between brain development and facial structure. However, the absence of distinguishable facial patterns in some genetic syndromes limits recognition methods based solely on facial traits.
Dingemans et al. introduced PhenoScore, an open-source, AI-based framework that quantifies phenotypic variations for rare genetic disorders without needing genomic data. PhenoScore is trained on facial features derived from 2D facial photos by a facial recognition model, combined with phenotypic similarity calculated based on the Human Phenotype Ontology (HPO) — a system that standardizes the description and analysis of phenotypic abnormalities found in human disease. Based on the combination of facial and phenotypic data, the framework predicts a score on how well the patient’s phenotype (as a whole) matches individuals with a known syndrome. PhenoScore identified unique phenotypes in 37 out of 40 investigated syndromes against clinical features observed in individuals with other neurodevelopmental disorders. This makes PhenoScore particularly useful in helping classify variants of uncertain significance.
Nature Genetics
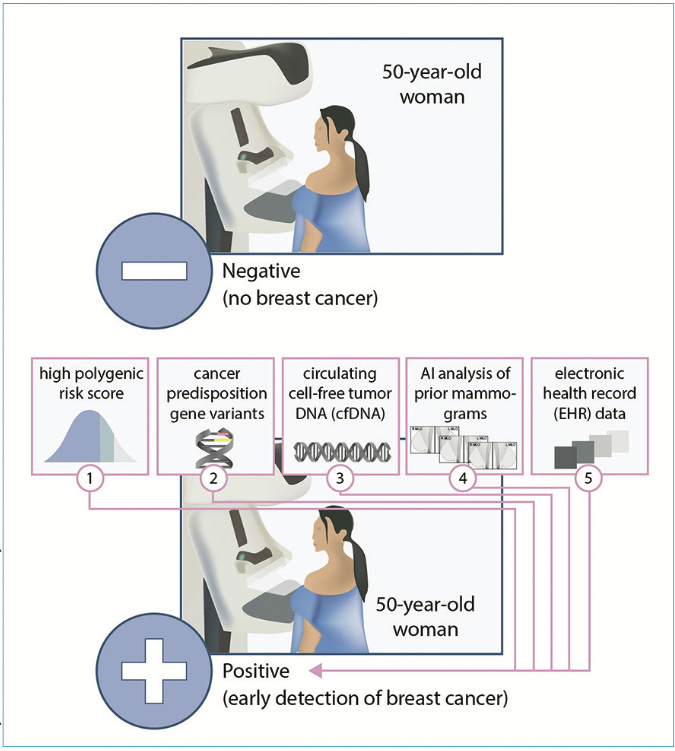
3. Cancer Screening. Current mass cancer screening methods, primarily based on age, are overly simplistic and can miss those who may be at high risk of disease.
Adams and Topol call for a revamp of cancer screening using AI. These AI models can harness newer data sources like genome sequencing and circulating cell-free tumor DNA (cfDNA). It can also extract information beyond human perception from routinely obtained medical images, such as X-rays, mammograms, and CT images. For instance, AI analysis of chest CTs can predict an individual's 6-year risk of lung cancer, which could be used to guide personalized screening intervals. By evaluating multimodal data sources, AI could introduce a "statistical biopsy," ushering in a more comprehensive and personalized strategy for early cancer detection.
The Lancet
4. Medical Education. Throughout history, technology has disrupted the way physicians think, and AI promises to influence the thought structures and practice patterns of medical trainees and physicians for years to come.
In this perspective, Cooper and Rodman emphasize the need to integrate AI into medical education. Medical trainees will have to become comfortable working with AI and understand its capabilities and limitations, not only to support their clinical skills but also because their patients are already using AI in their health inquiries. Such interactions will inevitably transform the patient–doctor relationship, mirroring how commercial genetic tests and online medical platforms previously changed discussion topics during clinic visits.
The New England Journal of Medicine
5. Medical Education. How can AI and data science be integrated into medical education?
In this Viewpoint article, Seth et al. propose methods for integrating AI and data science education into medical curricula. During the early pre-clerkship phase, as students begin interacting with clinical data through electronic health records, there is an opportunity to reinforce and delve deeper into topics related to data quality and coding. The authors suggest introducing students to the expanding role of AI in healthcare, exemplified by how Language Model Modules transform free text into structured data. Courses early in the pre-clerkship curriculum addressing medical ethics and professionalism also present an opportunity for an introduction to concepts in privacy and ethics of health data usage. In the clerkship phase, students can explore the use of data collected by consumer medical devices (such as step counters and sleep monitors) in clinical decisions. Likewise, students can engage in discussions about precision biologic treatments guided by genomic and proteomic screening, leveraging AI-generated personalized predictive analytics in light of the growing availability of "omics" data. Furthermore, throughout their clinical rotations, as students encounter real-world instances of data visualization and clinical decision support (CDS) tools, they can analyze the potential differential outcomes of machine learning algorithms due to inaccuracies or delays in capturing patient data, as well as shifts in patient demographics.
JMIR Medical Education
-- Emma Chen, Pranav Rajpurkar & Eric Topol